We are happy to announce the release of OntoMaton, a tool which allows users to search for ontology terms and tag free text right in Google Spreadsheets. This post will serve to introduce you to the tool, how it works and how it can make it easier for users to use ontologies in a pervasive, powerful and collaborative environment, complementing existing work from our team in the creation of ISAcreator.
How it looks
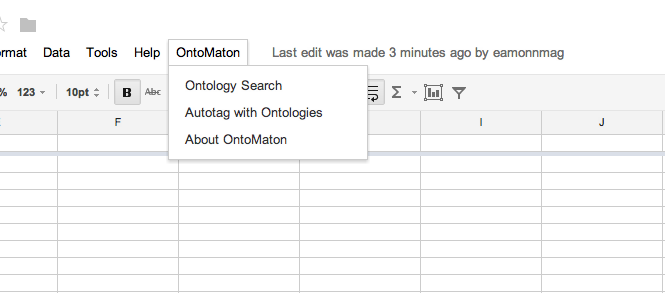
OntoMaton is available from the Google Script Gallery and when installed provides a menu as shown below.
From the menu you may access two resources part of OntoMaton: ontology search and ontology tagging. There is also an ‘about’ option.
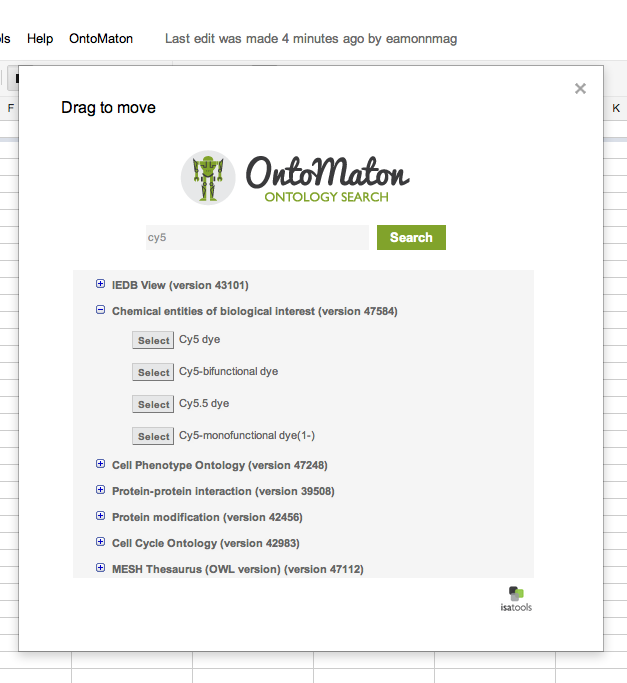
Ontology Search
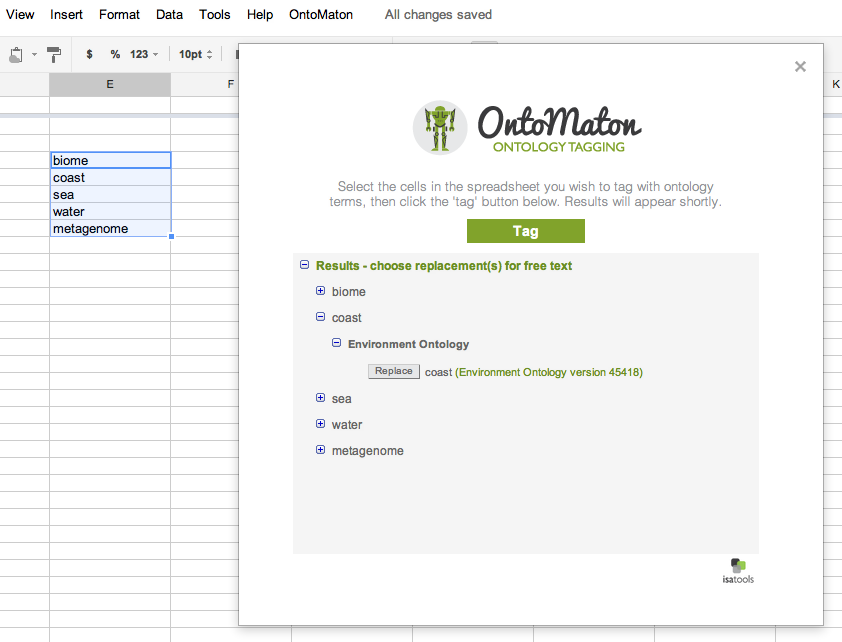
Ontology Tagging
Behind the scenes: restricting the ontology search space
If a sheet named “restrictions” is in your spreadsheet, OntoMaton will consult it to determine if the currently selected column/row name has a narrowed ontology search space. This makes it quicker to search BioPortal, allows for restriction of the user’s result space to make easier the process of selecting a term.
Behind the scenes: extra information about the terms you select
For every term you select, it’s full details are recorded in a “terms” sheet. This makes it possible to use OntoMaton in any spreadsheet and all provenance information (including URIs, ontology source and version) for selected ontology terms will be immediately available for use when exposing your records to the linked data world!
Installing
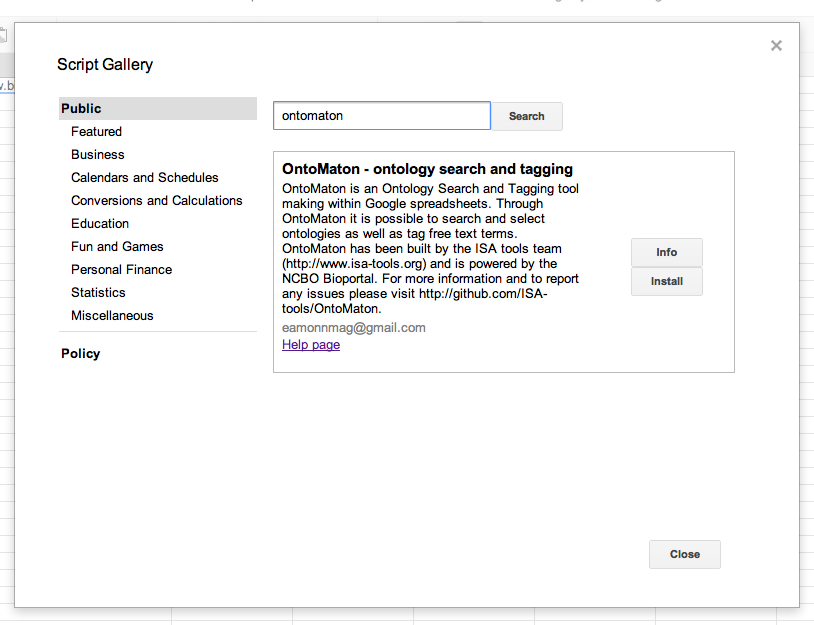
To install, create a new google spreadsheet, then go to the menu tools > script gallery. In the script gallery, search for ontology or ontomaton and you’ll get the following result pane.
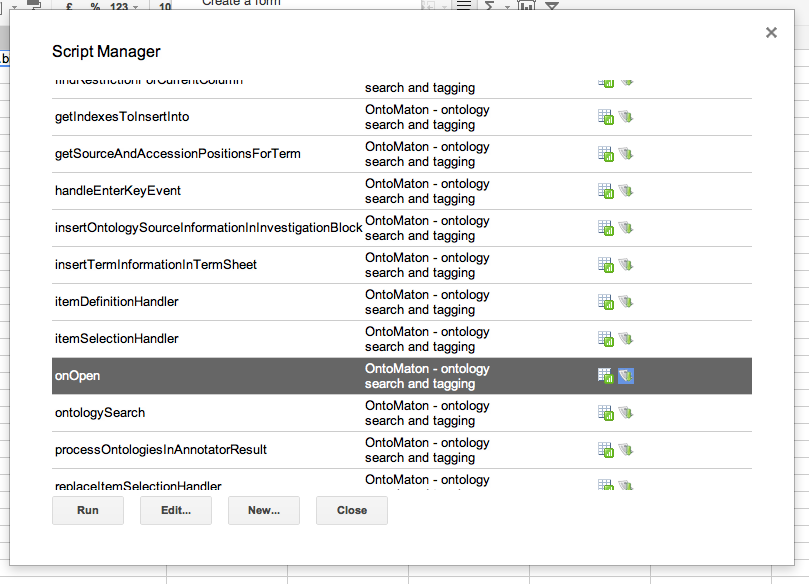
Click on ‘install’ and this will install the scripts inside your spreadsheet. Then there is one more and final step to follow for installation. You have to click again on tools > script manager and you’ll be presented with something like that shown in the image below.
OntoMaton contains lots of functions, but the only one you need to worry about in order to run the program is the onOpen function. Click this then click on run and the OntoMaton menu will be installed in your menu bar. From here you’ll be able to access the ontology search and ontology tagging functions.
Let us know what you think! New releases will come soon to fix any problems you may identify, please submit all ‘bugs’ and feature requests through https://github.com/ISA-tools/OntoMaton/issues
OntoMaton inherently supports ISA-Tab files too. So if you have an investigation file it will automatically add ontology sources to the ONTOLOGY SOURCE REFERENCE block. Also, if you have Term Source Ref and Term Source Accession after a column, OntoMaton will automatically populate these columns for you.
Also, the following table provides a quick review of available tools attempting to mix spreadsheets and access to vocabulary servers:
|
domain |
automated annotation |
ontology search/lookup |
versioning* |
collaboration |
|
|
RightField |
general |
✘ |
✓ |
✘ |
✘ |
|
ISA creator |
multiomics |
✓ |
✓ |
✘ |
✘ |
|
Proteome Harvest PRIDE |
proteomics |
✘ |
✓ |
✘ |
✘ |
|
Annotare |
transcriptomics |
✘ |
✘ |
✓ |
✘ |
|
OntoMaton |
general |
✓ |
✓ |
✓ |
✓ |
- by versioning we refer to managing of user edits throughout the annotation process.
We hope you enjoy this new feature!
The ISA team
Addendum:
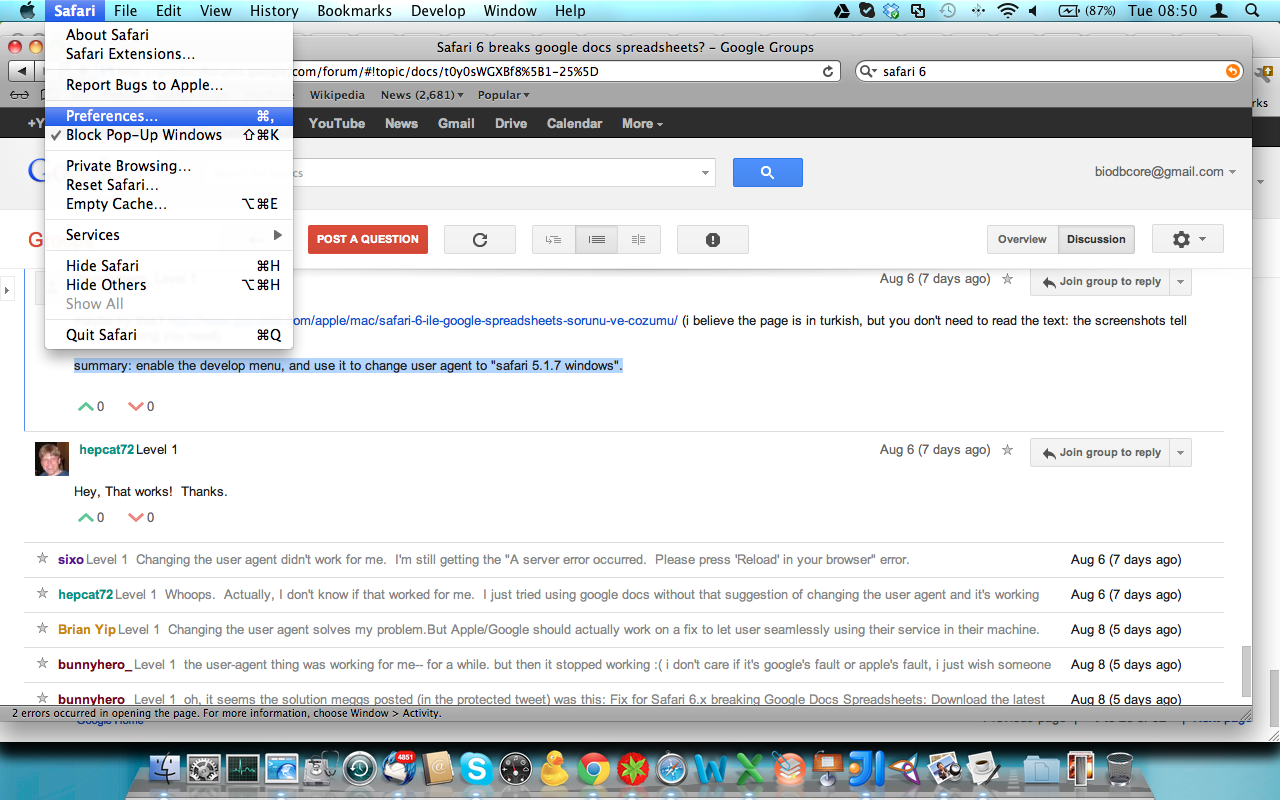
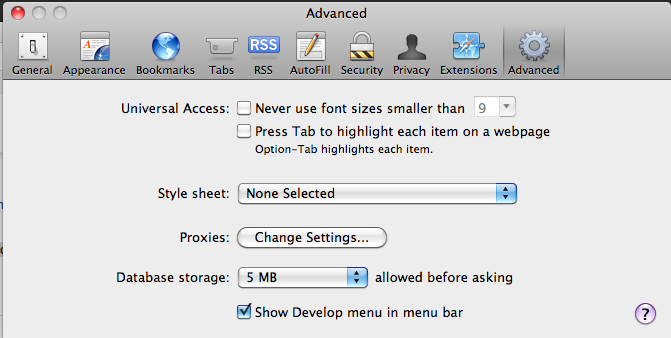
Safari 6 users, be aware you will have to activate the ‘developer menu’ from the Advanced Item in the Safari ‘Preferences’ menu item. Once activated, go to menu ‘Develop’ and navigate to ‘User Agent’ item and select ‘Safari 5.1.7’ for enabling the browser to work with Google Spreadsheet. (Thanks to rpyzh for reporting the issue, see here)