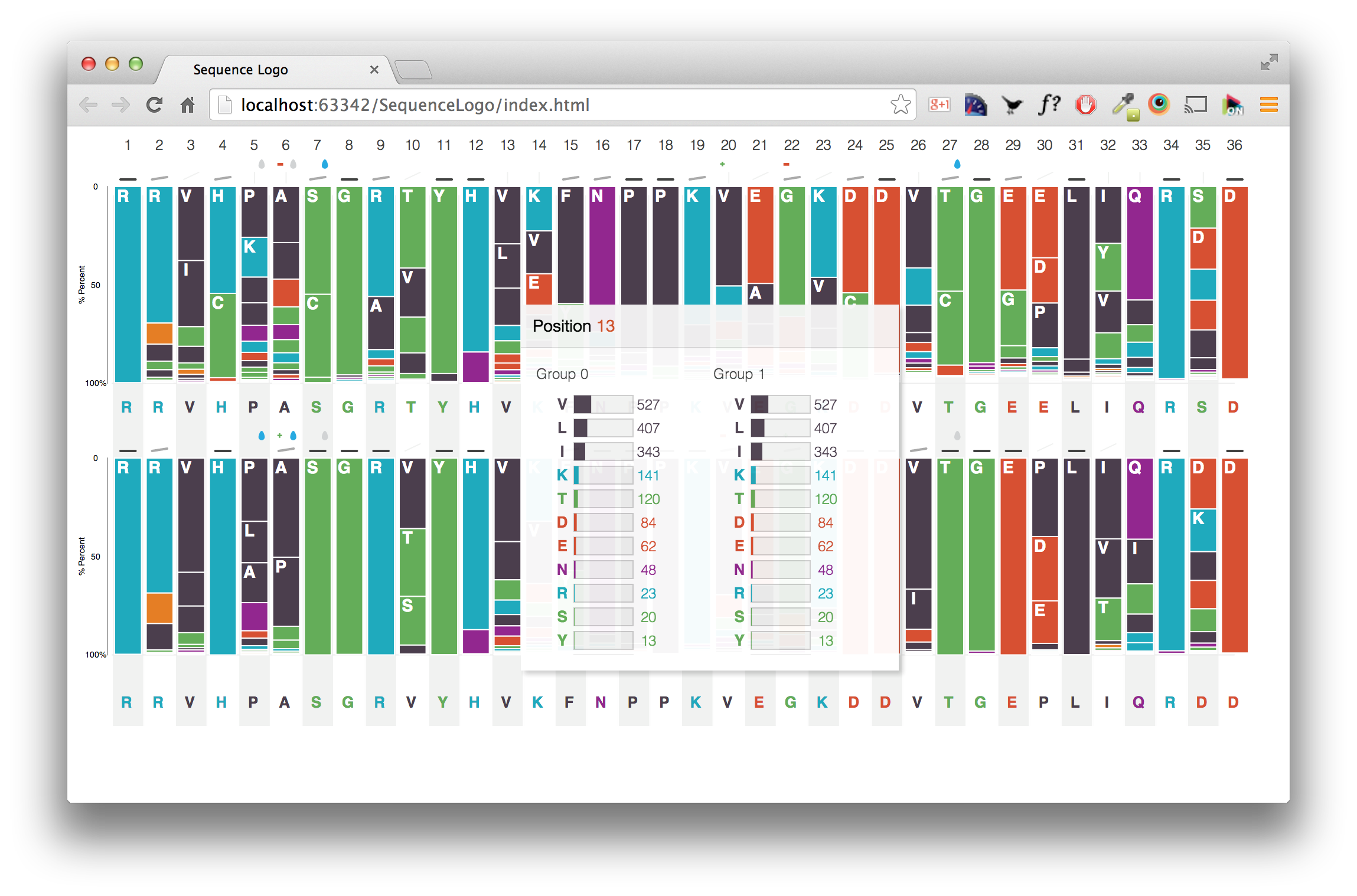
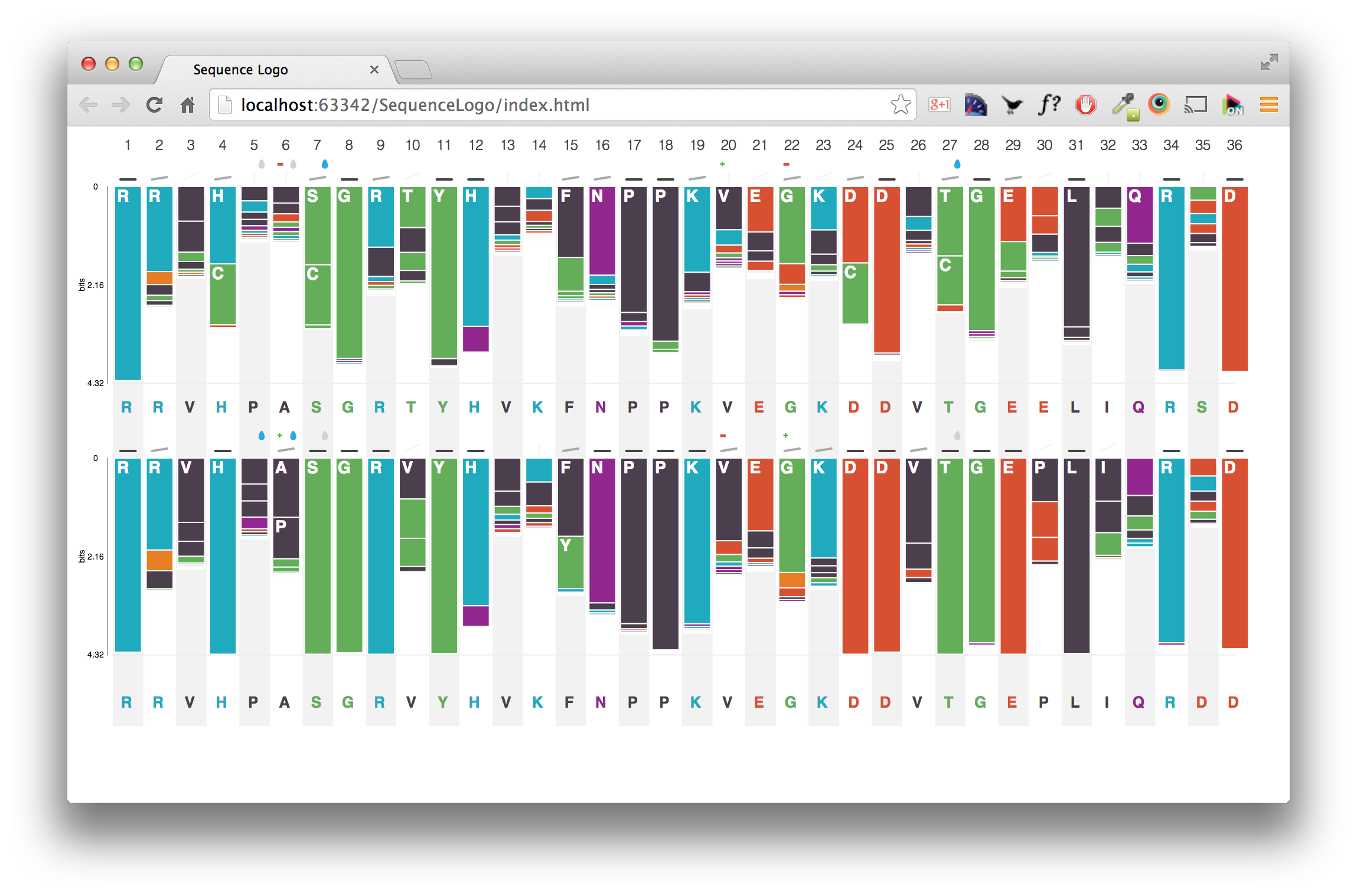
In true Star Wars fashion, we are bringing you the prequel to the last blog post discussing how to set up data file access from your JBoss installation. In this post we’ll discuss how to set up the BioInvestigation web application, from build to deployment.
1. Getting Ready
- First, you’ll need to have a Java installed.
- Then you’ll need to download Maven, this is what automates the build process for the BII web application. A bit like Ant, but it adds dependency management, meaning no need for pesky lib directories. Why this version I hear you ask? Well for some silly reason, the Maven developers (as great as they are) thought that removing the ability to specify profiles in profiles.xml was a great idea. We don’t think it is. In Maven 3+ you can only specify profiles in the pom.xml file or in the settings.xml file under the .m2 directory on your system. So, use version 2.2.1. We’ll move on to configuring these profiles a little later.
- Now you need Git which we’ll use to get the latest BII codebase from GitHub. If you haven’t heard of Git, you simply haven’t lived. Get out from underneath that SVN rock and join the Git revolution. It’s worth migrating just for the benefit of GitHub or some other funky equivalent (BitBucket or Launchpad).
- Next, download JBoss 5.1.0 from http://sourceforge.net/projects/jboss/files/JBoss/JBoss-5.1.0.GA and move the JBoss directory to somewhere to your liking. For further examples, let’s say that’s /etc/jboss-5.1.0.GA/ for the rest of this post.
- Finally, if you want to deploy properly, you’ll need a database. The BII works with Oracle, Postgres, MySQL or MariaDB. You can also run the application just for using the H2 database (we use this for testing). If your database of choice (apart from H2), you’ll need to create a database (we usually name it bioinvindex) and a user (it’s always a good idea to not just use root as the user) and grant that user all permissions to the database.
2. Grabbing the code
With everything in place, you need to ‘clone’ the BioInvIndex repository from GitHub. So navigate to a directory of your choosing, and via the command line type ‘git clone https://github.com/ISA-tools/BioInvIndex.git’. This will create a BioInvIndex directory for you with all the code inside.
3. Configure your profiles
I mentioned earlier about Maven and profiles. In the bioinvindex directory just created for you, you’ll see a profiles.xml file. Not to get too bogged down on details about what these profiles are for, I’ll give a brief explanation. The main pom.xml file (Project Object Model) for maven defines a number of dependencies, or libraries that are required for the application to run. Some of these dependencies are not always known in advance. For example, if I am running an application on MariaDB, why would I want to have Oracle and Postgres libraries in the final EAR (Enterprise Archive, the JBoss answer to WAR files in Tomcat)? The Profiles just allow us to automatically inject some properties in to the pom file on build depending on what the person building it wants to run. Some of these profiles also go towards creating the hibernate.properties file required by hibernate for connection to the database or in specifying to the application the whereabouts of the Lucene index (will talk more about that in a bit).
So now, given you have some context, open profiles.xml up and you’ll find a number of individual ‘profile’ elements for the various database installations and a few for the Lucene index locations.
First, find the profile matching your database and edit the username and password, database name (if not bioinvindex), URL and any version information, etc.
Then, modify the index profiles (named index_local and index_deploy) that specify where the Lucene index for the BII web app will reside. The Lucene index is used to speed up the web application, massively reducing the number of database queries that are required. The majority of the web application front end is based on the contents of the Lucene index. Make sure that the application has read/write access to that location.
4. Build!
Run the command mvn clean package -Dmaven.test.skip=true -Pdeploy,<your_database_profile>,<your_index_profile>
So, if I was deploying using mysql and using the index_deploy profiles, I would run
mvn clean package -Dmaven.test.skip=true -Pdeploy,mysql,index_deploy
Now watch the build succeed. When it completes, look in ear/target/ and you’ll see bii-x.y.ear where x and y are the major and minor versions respectively. The ear, or enterprise archive is what you’ll be deploying. Copy this file in to your JBoss directory, so given we’ve copied the JBoss installation files to /etc/jboss-5.1.0.GA/, we’d copy the ear to /etc/jboss-5.1.0.GA/server/default/deploy/
5. Run the web application
Finally, we want to actually run our application. The next step will rely solely on JBoss. navigate to /etc/jboss-5.1.0.GA/bin/ and you’ll see a number of scripts. The only one you need worry about is run.sh – to run JBoss using the default server (that’s where we copied the ear to), execute the following command – ./run.sh -c default -b 0.0.0.0 & the last bit will bind the server to address 0.0.0.0, so it’ll be visible to the outside world and will run on port 8080 by default. You can change the port by following the guidance in this Stackoverflow post.

Hopefully everything will start running, and the bii ear will be deployed and you’ll be able to see it at http://localhost:8080/bioinvindex. You will see errors in the startup log, but that’s because nothing exists in our database yet…there are no tables present at this point. We’ll create the database next.
6. Set up the database
For this, you’ll need the BII Manager. You can download this from http://isatab.sourceforge.net/tools.html. The latest version (1.6.1) is available here.
Unzip it and you’ll have a BII-Data-Manager-x.y directory with some files in it. We’ll need to configure some things before we start:
- Firstly, the database connection information. Open up the hibernate.properties file under the config/ directory and copy in the same username, password, database URL, index location, etc. as you entered in the profiles.xml for the BII web application.
- Next, you may want to set up data-locations.xml. Data locations tell the manager tool where to copy data files and ISA-Tab files as well as the URL the web application will be able to access them from. More information on the set up of JBoss to point to these data directories is available in a previous blog post.
Now, we’re ready to go. If you are using MySQL, H2, MariaDB or Postgres, you’ll be able to run the application by just double clicking isa_deps.jar or executing ./run.sh from the command line.
If you are using Oracle, you will need to add the Oracle driver on the classpath. Edit the run.sh file to do this then when ready, from the command line, run ./run.sh.
When the application starts, you’ll be presented with a log in screen. At this point, create a curator account. When this is done for the first time and the database has no schema available, the application will automatically create the schema via Hibernate (a fantastic tool). So you’ve nothing to do. Then, when that’s completed successfully (if it doesn’t check your hibernate.properties entries), you’ll be able to log in and load content.
Finally, to make things a bit faster, open up your hibernate.properties file having used the data manager for the first time and delete the line
hibernate.hbm2ddl.auto=update
This will stop the checking of the schema every time a connection is opened.
That’s it! Let us know if we missed anything, and we’ll update this post.