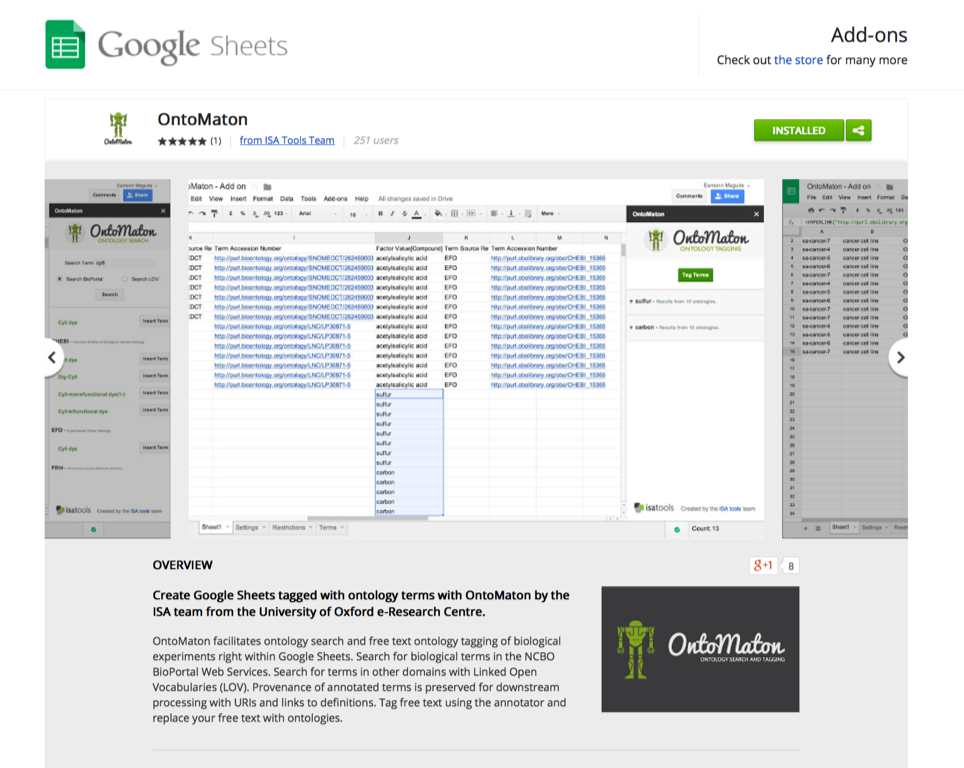
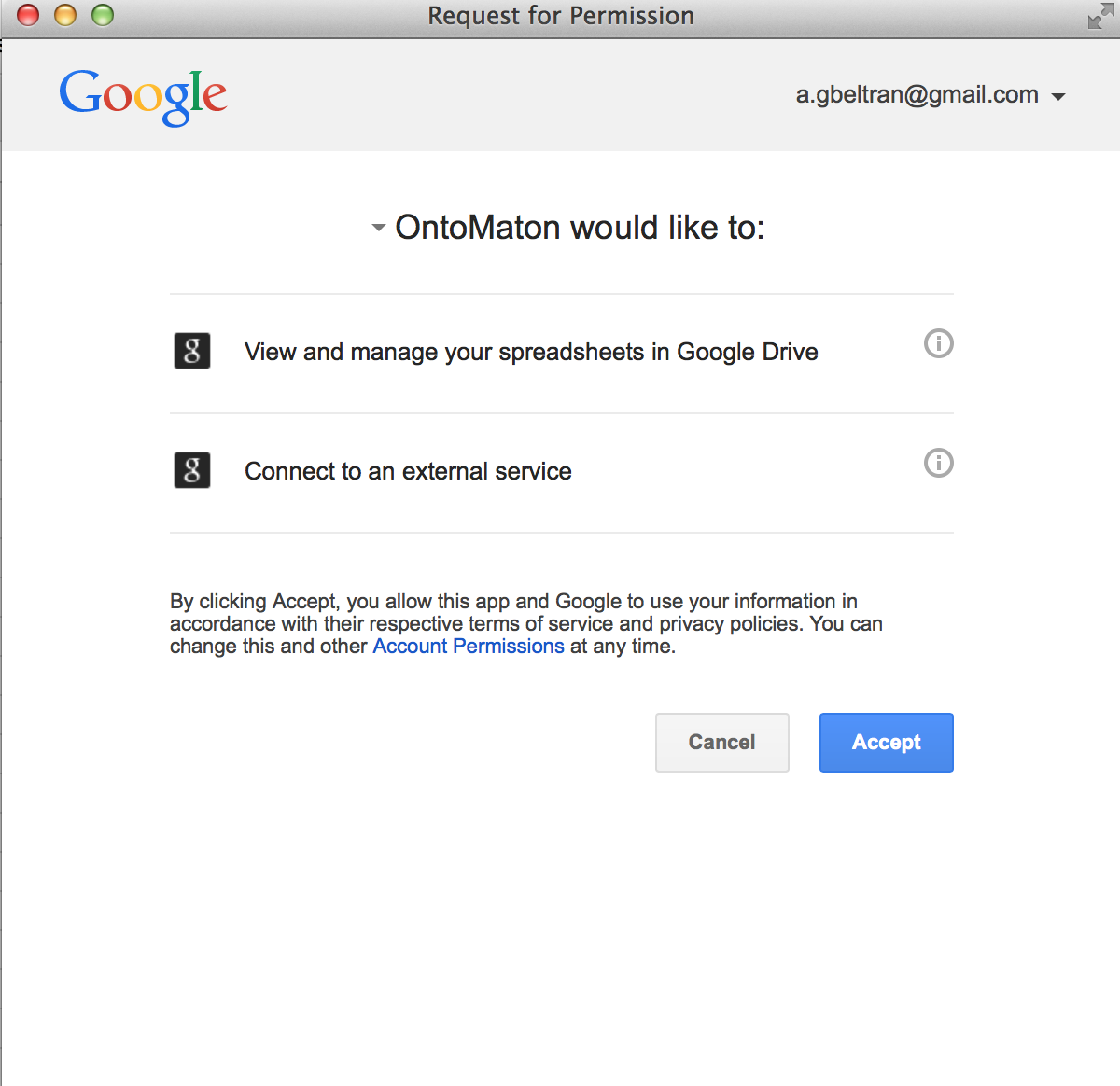
ISAcreator has supported access to GenomeSpace since release 1.7.0. and it is now also available through the GenomeSpace online interface.
GenomeSpace is a framework supporting cloud-based interoperability of genomics analysis tools. By providing access to multiple tools through their interface, and supporting file transfers in the cloud, GenomeSpace provides a bridge among the tools, allowing users Some of the tools available through GenomeSpace are: Cytoscape, Galaxy, GenePattern, Genomica, Integrative Genomics Viewer (IGV), and the UCSC Genome Table Browser. Find out more about what is GenomeSpace and what GenomeSpace can do for you.
I will now describe the functionality ISAcreator supports for GenomeSpace.
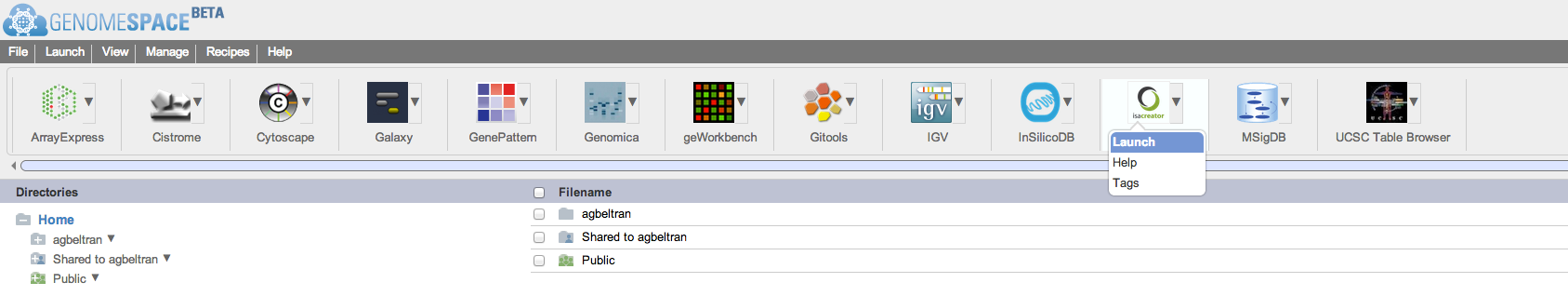
You can launch ISAcreator from your desktop or you can launch it once you are logged in to GenomeSpace (after registering to their service). For launching ISAcreator from within GenomeSpace, just hover over the ISAcreator icon and select ‘Launch’:
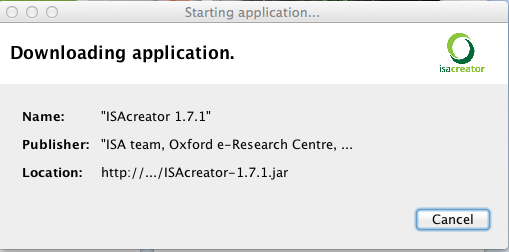
If you launch ISAcreator from GenomeSpace, you will be prompted to download ISAcreator and will see the following pop-up window (after you accept to download the file):
When running ISAcreator (either from your desktop or following the GenomeSpace route), you will notice that it now has a third mode of operation (apart from the previously available light and normal modes) that corresponds to GenomeSpace. With this third method of operation, ISAcreator supports opening ISA-TAB files stored on the cloud environment provided by GenomeSpace and also, saving files into GenomeSpace storage facilities.

If you choose the GenomeSpace mode, you will have to enter your GS user credentials in the ISAcreator login page:

Then you will load the configuration files, as usual, and get to the main menu where you can choose to load an existing ISA-TAB file. If it is not the first time you are loading files, you will see the previously loaded files and also have the option to search GenomeSpace for more files:

As an example, you can find the publicly available BII-I-1 ISA-TAB dataset in GenomeSpace under Public/agbeltran/ISA-TAB-datasets, and select it to load:
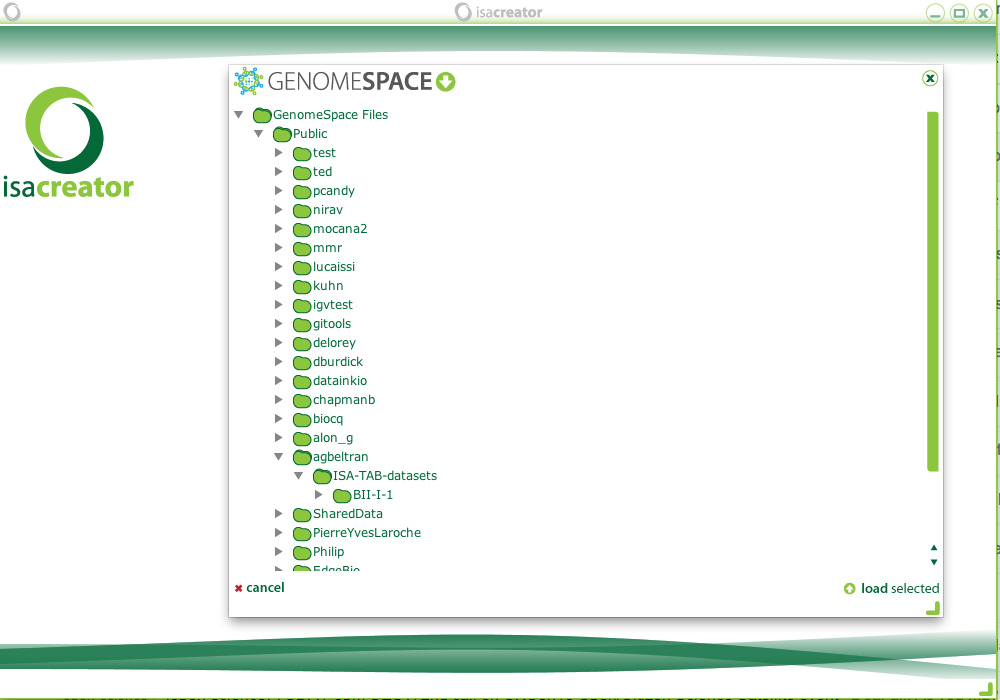
After loading an ISA-TAB dataset, you can save it to GenomeSpace (even if it is a local dataset that you want to store in GenomeSpace):
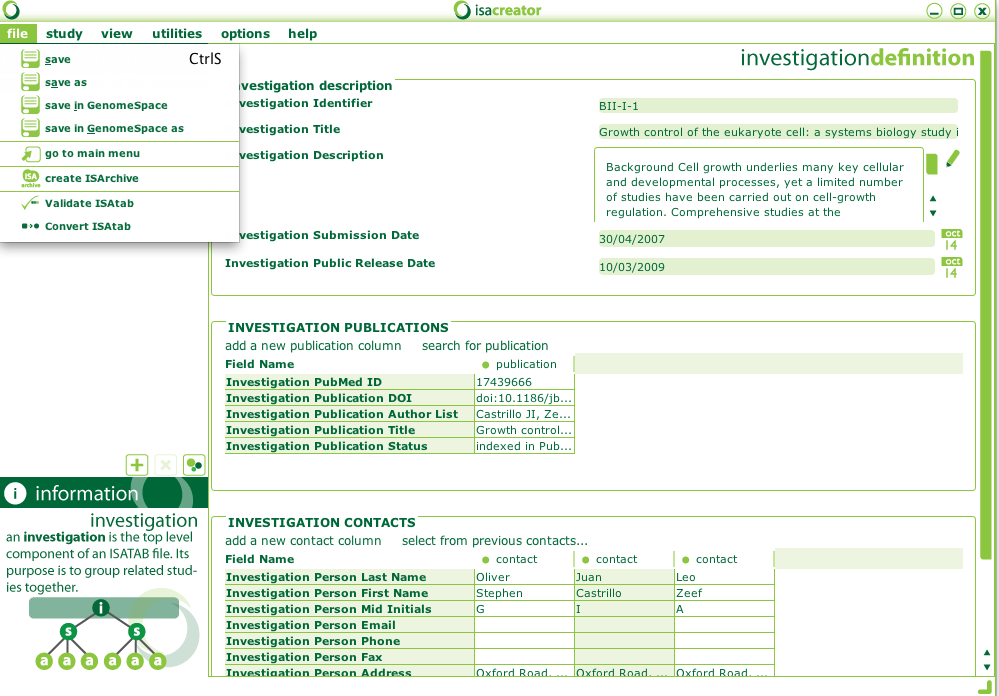
GenomeSpace also provides documentation about ISAcreator in this page and a guide about using ISAcreator in this other page.
As always, send us comments or questions contacting:
- the ISA team at isatools [at] googlegroups [dot] com,
- the ISA user forum at isaforum [at] googlegroups [dot] com
or send us feature requests or bug reports through the issue tracker in Github:
- https://github.com/ISA-tools/ISAcreator/issues